What’s the advantages of using HDF5 for file saving and loading?
I wrote something about pickle or JSON before, which are python packages for serialization.
More specifically, pickle is a binary serialization format for python objects, saving objects to an unreadable file, can be loaded inside the same machine and is not sharable with other programming languages.
And JSON is a text serialization which saves basically python dictionaries, text, list like object in a readable format. And it’s sharable, and not limited to python language.
Another saving option is CSV, which often related python package pandas. Pandas can read csv files into pandas data frames, or save data frames to csv files easily, and further manipulate them. CSV is a format that’s perfect for tabular data, can be readily shared to a wide range of other software.
And then we have .npy as well as .npz format files which are supported by numpy methods for saving of one (.npy) or multiple compressed (.npz) numpy arrays. np.save and np.savez functions are for saving and np.load is for loading.
But basically none of the above, can nicely save/load a bunch of numpy array as images, with their meta-data, which I think is basically where HDF5 files comes in.
Structure of HDF5 files
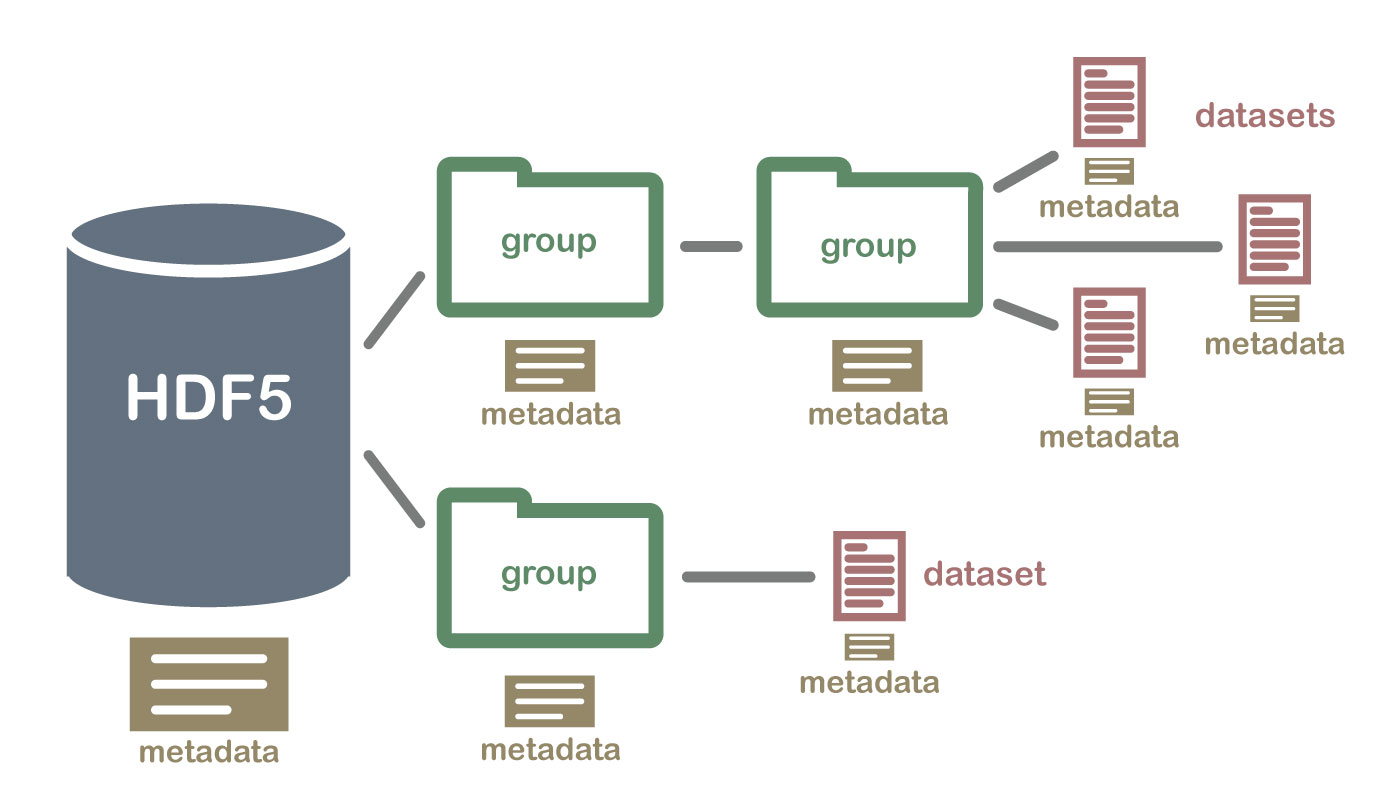
There are groups and datasets inside a HDF5 file, we can think of groups as folders, datasets as files. And for each folder and file, we can have descriptions/tags/metadata for them.
Groups
Groups are the container mechanism by which HDF5 files are organized. From a Python perspective, they operate somewhat like dictionaries. In this case the “keys” are the names of group members, and the “values” are the members themselves (
GroupandDataset) objects.
Datasets
Datasets are very similar to NumPy arrays. They are homogeneous collections of data elements, with an immutable datatype and (hyper)rectangular shape. Unlike NumPy arrays, they support a variety of transparent storage features such as compression, error-detection, and chunked I/O.
They are represented in h5py by a thin proxy class which supports familiar NumPy operations like slicing, along with a variety of descriptive attributes:
- shape attribute
- size attribute
- ndim attribute
- dtype attribute
- nbytes attribute
HDF5 for python: h5py
h5py is a python package for hdf5 file processing, here are some most basic usage of this package
Opening & creating files
>>> f = h5py.File('myfile.hdf5','r') # 'r' for reading
File mode list:
| Mode | Description |
|---|---|
| r | Readonly, file must exist (default) |
| r+ | Read/write, file must exist |
| w | Create file, truncate if exists |
| w- or x | Create file, fail if exists |
| a | Read/write if exists, create otherwise |
Creating groups
>>> grp = f.create_group("bar")
>>> grp.name
'/bar'
>>> subgrp = grp.create_group("baz")
>>> subgrp.name
'/bar/baz'
Comment: just like a file system.
Creating datasets
>>> dset = grp.create_dataset("default", (100,))
>>> dset = grp.create_dataset("ints", (100,), dtype='i8')
create_dataset ( name**,** shape=None**,** dtype=None**,** data=None**,** **kwds )
Once the shape is specified, we can write data using slices, indices …
Attributes
Meta-data for groups and datasets: accessible by using group.attrs or dataset.attrs, like a python dictionary:
keys()Get the names of all attributes attached to this object.Returns:set-like object.
values()Get the values of all attributes attached to this object.Returns:collection or bag-like object.
items()Get
(name, value)tuples for all attributes attached to this object.Returns:collection or set-like object.get(name, default=None)Retrieve name, or default if no such attribute exists.
get_id(name)Get the low-level
AttrIDfor the named attribute.create(name, data, shape=None, dtype=None)Create a new attribute, with control over the shape and type. Any existing attribute will be overwritten.
- Parameters:
- name (String) – Name of the new attribute
- data – Value of the attribute; will be put through
numpy.array(data). - shape (Tuple) – Shape of the attribute. Overrides
data.shapeif both are given, in which case the total number of points must be unchanged. - dtype (NumPy dtype) – Data type for the attribute. Overrides
data.dtypeif both are given.
- Parameters:
Potential problems of HDF5
One thing to be concerned about is that when your hdf5 file is supper large, loading all of them into memory is a bit not efficient, especially when your code only needs a small portion of the data from a large file, constantly…
An alternative when you are in this kind of situation is to make it the code to be able to load only part of the data but not whole. And that indicates we store the data into smaller chunks which when we only need part of them each time.
How to do that? Well, very simple, just store your data as a file system! Previously: group1/sub_group2/dataset1/, now folder1/sub_folder2/array1. For arrays, we can use numpy saving option: .npy files. And for meta-data, it’s a bit tricky here.
Saving metadata. One easy way to do it is to use a text serialization file format, e.g. JSON. But since JSON is not python specific, there are some classes in python that json cannot recognize, e.g. Tuples. JSON file will interpret tuples as the same as list, which is something to keep in mind when you want to store a dictionary that contains tuples…